Marker enrichment and fine mapping of the barley 5H(7) chromosomal region homologous to the Ph1 gene region of wheat
Mohammad M. Shah and Kulvinder S. Gill
Department of Agronomy, University of Nebraska-Lincoln
P. O. Box 9015, NE 68583, USA
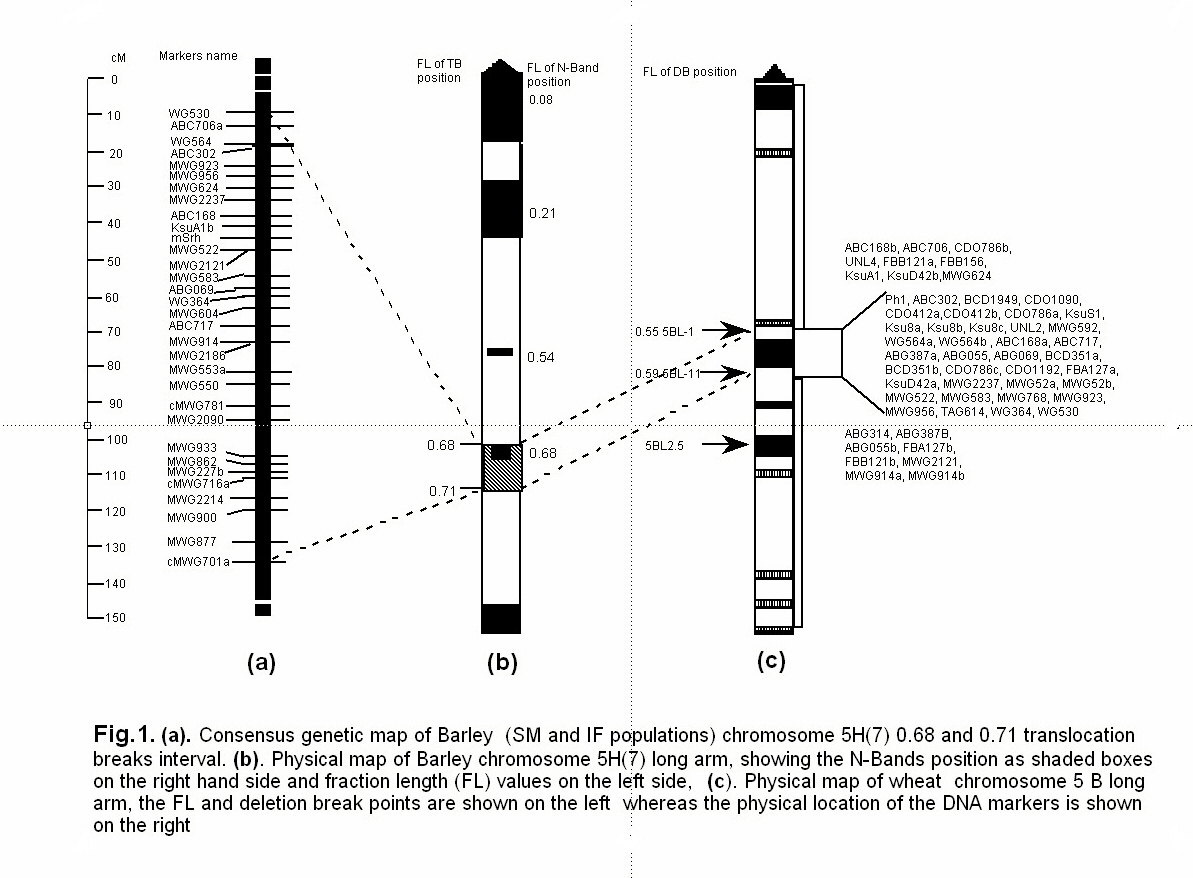
The region present between fraction length (FL) 0.68 and 0.71 of the long arm of chromosome 5H of barley contains a major gene cluster and is a recombination hot spot (Kunzel et al., 2000). Nineteen of the 42 markers on the Igri X Franka (I/F) 5H genetic map (Graner et al., 1991, Graner et al., 1993) are present in the region (Kunzel et al., 2000). The recombination is very high and the Mb/cM ratio in the region ranges from 0.1 to 0.6. Part of this region was identified to be homologous to the Ph1 gene region of wheat via commonly mapped markers (Gill et al., 1996a; Faris et al., 2000). The Ph1, which is a principal gene regulating chromosome pairing in wheat, has been precisely mapped to a submicroscopic chromosomal region on the wheat chromosome 5BL, spanning less than 1Mb (Gill et al. 1991; Gill et al. 1993). In wheat the region is bracketed by the breakpoints of single-break deletion line 5BL-1 on the proximal end and 5BL-11 at the distal end. This region will be defined to as the ‘Ph1 gene region’. In spite of its precise mapping in wheat, molecular characterization of the Ph1 gene in barley would be more cost effective because of the simpler system and the availability of better resources. Therefore our primary objective is to target the barley homologue of the Ph1 gene. The other possible targets in the gene-rich region (FL 0.68 and 0.71) on 5H of barley are short rachilla hair (Srh), high lysine (Lys1), seed width, (-amylase activity, (-glucanase activity, free as well as total limit dextrinase activity, compositum (Com1), Eceriferum (Cer), erectoides (Ert-g and Ert-n), and breviaristatum (Ari-e).
By comparative mapping of the previously mapped RFLP probes in the Ph1 gene region (Gill et al., 1993b; Gill et al., 1996a) with various maps in the Triticeae (such as wheat, barley, oat, rice, T. tauschii and T. monococum) we identified 52 potential probes for the region. Additionally, 10 potential markers were identified for the region using RNA fingerprinting/Differential display (RNAF/DD). Forty-five out of the total 62 markers were tested by gel blot DNA hybridization analysis on the blots containing HindIII and DraI or EcoRI digested DNA of the three nullisomic-tetrasomic for wheat homoeologous group 5 chromosomes [NT5A(5B), NT5B(5A), NT5D(5A)], Ph1b mutant and the single-break deletion lines 5BL-1 and 5BL-11, as previously described (Gill et al., 1993b). The Ph1b mutant line was used to specifically assign the 5BL fragment band to the submicroscopic interstitial deletion encompassing the Ph1 gene. Probes detecting a 5BL chromosome specific fragment band that was absent in 5BL-1 but present in 5BL-11 were localized to the Ph1 gene region. The 5B specific fragment bands present in both of the deletion lines 5BL-1 and 5BL-11 were mapped to the proximal end of the deletion breakpoint of 5BL-1, whereas the fragment bands absent in these deletion lines were mapped distal to the deletion breakpoint of 5BL-11. The Ph1 gene region probes identified by this approach, were genetically mapped using S/M barley population. Ten of the region probes were previously mapped on the same S/M barley population (Kleinhofs et al., 1993) and 5 on the I/F population (Graner et al., 1991). The two genetic maps (i.e. S/M and I/F) of the 5H chromosome of barley corresponding to the Ph1 gene homologue region between 0.68 and 0.71 translocation intervals (Kunzel et al., 2000) were combined to generate a consensus genetic map (Fig. 1. a.). The physical map of the 5H long arm of barley was reproduced from Kunzel et al. (2000) (Fig.1. b), whereas the physical map of 5BL of wheat for the Ph1 gene region was drawn as described by Gill et al. (1993b). The Ph1 gene region is marked by the dotted lines across the maps (Fig. 1. a, b, c).
Forty potential Ph1 gene region probes detected 49 loci for chromosome 5B of wheat. Five of the probes failed to detect any locus for homoeologous group 5. Thirty-seven loci mapped in the Ph1 gene region (Fig.1. c). Nine loci present in both deletion lines 5BL-1 and 5BL-11, therefore map proximal to the Ph1 gene region. Similarly 8 probes mapped distal to the Ph1 gene region. Five probes were found commonly mapped between I/F and S/M 5H barley genetic maps, which were previously mapped in translocation intervals 0.68 and 0.71 (Kunzel et al., 2000). Three of these 5 probes were mapped in the Ph1 gene region with 2 mapped distal to it. The conserved region between barley and wheat showed high recombination (Fig.1.) and the order of the markers on both of the genetic maps of barley were the same. All the probes identified for the homologous Ph1 gene region are being used to screen 1.5X barley BAC library and the positive BAC clones will be assembled into a contiguous map of the region, for eventual targeting the useful genes present in the region.
Faris, J. D., K. M. Haen, and B. S. Gill. 2000. Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics 154: 823-835
Gill, K.S., B.S. Gill, and T.R. Endo. 1993a. A chromosome region-specific mapping strategy reveals gene-rich telometric ends in wheat. Chromosoma 102:374-381.
Gill, K.S., B.S. Gill, T.R. Endo, and Y. Mukai. 1993b. Fine physical mapping of Ph1, a chromosome pairing regulator gene in polyploid wheat. Genetics 134:1231-1236.
Gill, K.S., B.S. Gill, T.R. Endo, and E. Boyko. 1996a. Identification and high-density mapping of gene-rich regions in chromosome group 5 of wheat. Genetics 143:1001-1012.
Graner, A., E. Bauer, A. Kellermann, S. Kirchner, and J.K. Muraya. 1993. Progress of RFLP-map construction in winter barley. Barley Genet. Newslett. 23:53-59.
Graner, A., A. Jahoor, J. Schondelmaier, H. Siedler, K. Pillen, G. Fischbeck, G. Wenzel, and R.G. Herrmann. 1991. Construction of an RFLP map of barley. Theor. Appl. Genet. 83:250-256.
Kleinhofs, A., A. Kilian, M.A. Saghai Maroof, R.M. Biyashev, P. Hayes, F.Q. Chen, N. Lapitan, A. Fenwick, T.K. Blake, E. Anaiev, L. Dahleen, D. Kudrna, J. Bollinger, S.J. Knapp, B. Liu, M. Sorrells, M. Heun, J.D. Franckowiak, D. Hoffman, R. Skadsen, and B.J. Steffenson. 1993. A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor. Appl. Genet. 86:705-712.
Kunzel, G., L. Korzum, and A. Meister. 2000. Cytologically Integrated Physical Restriction Fragement Length Polymorphism Maps for the Barley Genome Based on Translocation Breakpoints. Genetics 154:397-412.