In connection with localization of QTL a linkage map (including chromosome 5) of mainly molecular and biochemical markers were presented (Thomas et al. 1995a, 1995b). The map was based on DH-lines from a cross between variety 'Blenheim' and a breeding line 'E224/3'.
QTL for beta-glucan and beta-glucanase activity in grain and malt were associated chromosome 5 in the 'Stepto' x 'Morex' cross (Han et al. 1995).
A new type of molecular markers RAMPs/dRAMPs were reported (Becker and Heun 1995). The markers were studied in the 'Proctor' x 'Nudinka' cross (Heun et al. 1991). Forty different loci were reported. They were distributed on all 7 barley chromosomes and were mapped on the previously reported 'Proctor' x 'Nudinka' map (Heun et al. 1991). Becker and Heun (1995) report that the new loci cause stretching of the linkage map. They suggest it is because of specific localized recombination hot spots. The most common reason for stretching of the linkage maps, incorrectly classified genotypes, was not discussed.
A linkage map mainly based on RFLP markers and DH-lines from the cross 'Tystofte Prentice' x 'Vogelsanger Gold' was reported in connection with a QTL study (Kjær et al. 1996).
ADP-glucose pyrophosphorylase genes Aga7 and Aga6 were located on respectively the minus and the plus arm of chromosome 5 by means of wheat-barley telosomic addition lines (Kilian et al. 1994).
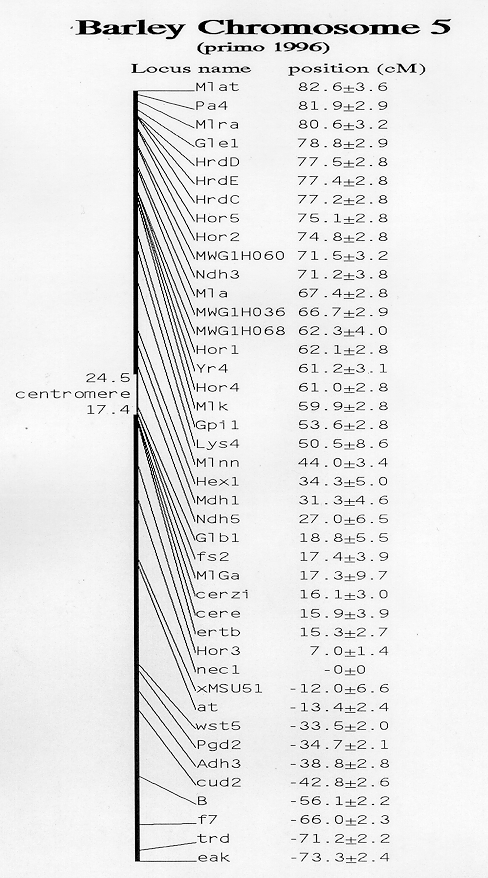
Using microisolated translocated chromosomes it was shown that the RFLP marker MWG913 is located on the plus arm of barley chromosome 5 (Sorokin et al. 1994, 1995). Further, the marker was localized 32.5 cM proximal to Hor1 on the linkage map of Graner et al. (1991). Thus according to Figure 1 the centromere may be in position lower then 62.1-32.5=29.1 cM. This is not in conflict with the upper bound of the centromere position suggested in last year's coordinator report based on data from Schondelmaier et al. (1993). The reports of Sorokin et al. (1994, 1995) further confirm that recombination is physically localized distally on chromosome 5.
A consensus linkage map was constructed based mainly on RFLP markers from seven crosses (Langridge et al. 1995). Linkage maps from three of the crosses have previously been published ('Igri' x 'Franka', 'Proctor' x 'Nudinka' and 'Stepto' x 'Morex'). The linkage data of the other four crosses, made at Waite Institute, Adelaide, has apparently not been published before. The consensus map of chromosome 5 includes 77 marker loci and is 172 cM long. Five loci (Hor5, Hor2, Mla6, Hor1 and Pgd2) are common with loci on the map in Fig 1. The distance between those loci on the consensus map are respectively, 0.7, 7.4, 2.3 and 94,5 cM. On the map in Figure 1 the corresponding distances are 0.3±0.23, 7.5±0.49, 5.2±0.45 and 96.8±3.05 cM. The only distance that deviate significantly is between Mla6 and Hor1. However, Mla6 may be different from Mla in Figure 1 (see co-ordinators report BGN vol. 24). By comparing the two maps it can be seen that the map in Figure 1 is 40 cM shorter on the plus arm and 18 cM longer on the minus arm, than those on the consensus map.
None of the above linkage data were included in calculating the linkage map of chromosome 5 shown in Figure 1. However, if the data used for estimation the consensus map (Langridge et al. 1995) were included then the map would have had 114 loci instead of 42, and the map would have been 195 cM long instead of 155 cM. Furthermore, the map positions would have been more precise, although not much because of the few common markers.
The linkage map shown in Figure 1 is calculated based on linkage information reported in the former issues of BGN and on the procedure for estimating linkage maps by Jensen (1987). In addition is used the recombination percentages of 12±2 reported between Hor1 and Hor2 (Kapala and Patryna 1993). This value has not changed the map estimates. The only reason why the map in Figure 1 is different from the map in last years report (BGN vol. 24) is that I have made a misprint in the map of chromosome 5.
References:
Becker, J. and M. Heun. 1995: Mapping of digested and undigested random amplified microsatellite polymorphisms in barley. -Genome 38:991-998.
Graner, A., A. Jahoor, J. Schondelmaier, H. Siedler, K. Pillen, G. Fischbeck, G. Wenzel and R.G. Herrmann. 1991: Constrution of an RFLP map of barley. -Theor. Appl. Genet. 83:250-256.
Han, F., S.E. Ullrich, S. Chirat, S. Menteur, L. Jestin, A. Sarrafi, P.M. Hayes, B.L. Jones, T.K. Blake, D.M. Wesenberg, A. Kleinhofs and A. Kilian. 1995: Mapping of beta-glucan content and beta-glucanase activity loci in barley grain and malt. -Theor. Appl. Genet. 91:921-927.
Heun, M, A.E. Kennedy, J.A. Anderson, N.L.V. Lapitan, M.E. Sorrells and S.D. Tanksley. 1991: Construction of a restriction fragment length polymorphism map for barley (Hordeum vulgare). -Genome 34:437-447.
Jensen, J. 1987: Linkage map of barley chromosome 4: -In: Barley Genetics V. Okayama, Japan:189-199.
Kapala, A. and H. Patyna. 1993: Genetic control of hordein polypeptides in kernels of spring barley (Hordeum vulgare L.) high-lysine mutant C-67-7. -Genetica Polonica 34:133-138.
Kilian, A., A. Kleinhofs, P. Villand, T. Thorbjørnsen, O.-A. Olsen and L.A. Kleczkowski. 1994: Mapping of the ADP-glucose pyrophosphorylase genes in barley. -Theor. Appl. Genet. 87:869-871.
Kjær, B., J. Jensen and H. Giese. 1996: Quantitative trait loci for heading date and straw characters in barley. -Genome (in press).
Langridge, P., A. Karakousis, N. Collins, J. Kretschmer and S. Manning. 1995: A consensus linkage map of barley. -Molecular Breeding 1:389-395.
Schondelmaier, J., R. Martin, A. Jahoor, A. Houben, A. Graner, H.-U. Koop, R.G. Herrmann and C. Jung. 1963: Microdissection and microcloning of the barley (Hordeum vulgare L.) chromosome 1HS. -Theor. Appl. Genet. 86:629-636.
Sorokin, A., F. Marthe, A. Houben, U. Pich, A. Graner and G. Knzel. 1994: Poly- merase chain reaction meditated localization of RFLP clones to microisolated trans- location chromosomes of barley. -Genome 37:550-555.
Sorokin, A., F. Marthe and G. Künzel. 1995: Integration of 37 translocation breakpoints of barley chromosome 5 into the Igri/Franka derived RFLP map. -BGN 24: 108-112.
Thomas, W.T.B., W. Powell, R. Waugh, K.J. Chalmers, U.M. Barua, P. Jack, V. Lea, B.P. Forster, J.S. Swanston, R.P. Ellis, P.R. Hanson, and R.V.M. Lance. 1995a: Detection of quantitative trait loci for agronomic, yield, grain and disease characters in spring barley (Hordeum vulgare L.). -Theor. Appl. Genet. 91:1037-1047.
Thomas, W.T.B., W. Powell, R. Waugh, B.P. Forster, K.J. Chalmers, U.M. Barua, J.S. Swanston, R.P. Ellis, P. Jack and V. Lea. 1995b: Quantitative trait loci in a North West European barley cross-Blendheim x E224/3. -BGN.24.41-45.
Wang , S. 1995: Association of the chlorina gene f3 with chromosome arm 5S in barley. -J. Heredity 86:151-152.
Legend to figure
Fig. 1. The barley chromosome 5 linkage map with the best fit to all linkage data available in the scientific literature calculated according to Jensen (1987). The map positions are given in centimorgans (cM). The order and positions of closely linked loci may not be definitive.
